Comparative and Semi-Quantitative Proteomics Using 2D Gel Electrophoresis (2D-DIGE) Technology
ITSIBIO »ITSI-Biosciences utilizes validated sample preparation kits/protocols, robotic spot pickers/digesters, DeCyder difference-in-gel analysis software and advanced mass spectrometers to offer an integrated 2D-DIGE – MS/MS proteomics solution. Proteomic analysis usually starts from sample preparation, to analyses by mass spectrometry (LC-MS/MS) and protein identification and annotation. Data obtained after 2D-DIGE analysis may include protein identity, pI, molecular weight and post translational modifications.
ITSI-Biosciences has provided 2D-DIGE analysis and proteomics services to NIH, industry, biotechnology companies, pharmaceutical companies, Research Institutes and Universities. Our more than 15 years of experience with 2D-DIGE analysis, proteomics, mass spectrometry and protein expression profiling enables us to offer reproducible picture perfect gels at a competitive price and guaranteed faster turnaround time. We have also developed a variety of complementary 2D-DIGE kits to streamline and standardize protein isolation, protein quantitation, protein qualification and the 2D-DIGE analysis step, thereby reducing the overall time and cost of performing 2D-DIGE analysis.
2D-DIGE offers all the advantages of the classical 2D-PAGE, and most importantly overcomes the inherent disadvantage of gel-to-gel variation and reproducibility problem inherent with the traditional 2D-PAGE approach. 2D-DIGE is presently the most versatile gel-based approach for comparative and semi-quantitative proteomics. Distinct fluorescent tags e.g. Cy 3, 5 and 2 are used to label protein samples and a universal internal standard prior to 1st/2nd dimension electrophoresis. An automated software program is used to detect, quantify and annotate differentially expressed proteins. 2D-DIGE analysis can be applied to protein expression profiling and comparative proteomics by using DeCyder software to identify proteins with statistically-significant difference in expression. This provides unique opportunities for researches in fields including cancer, neuroscience, vision, cardiovascular, Alzheimer, Parkinson, etc, to find important proteins associated with the condition. 2D-DIGE – LC/MS/MS analysis can also be applied to identify post translationally modified proteins (PTM) that are important in cell signaling and protein trafficking.
| S/N | CAT NO. | NAME | DESCRIPTION | UNIT COST |
|---|---|---|---|---|
| 1 | SP1 | Sample Preparation 1 | Homogenization, protein quantitation using ToPA Bradford protein assay kit. | Per sample |
| 2 | SP2 | Sample Preparation 2 | Includes, precipitation, buffer exchange, protein quantitation, etc. and the use of ToPI-DIGE, ToPREP and ToPA kits. | Per sample |
| 3 | SP3 | Sample Preparation 3 | Depletion of abundant protein e.g. bovine or human serum albumin using ITSI ASK-Column kit. | Per sample |
| 4 | SP4 | Sample Preparation 4 | Depletion of abundant protein e.g. bovine or human serum albumin using ITSI ASK-Solvent kit. | Per sample |
| 5 | SP5 | Sample Preparation 5 | Depletion IgY Column Depletion of 14 abundant proteins from serum /plasma using IgY column. | Per sample |
| 6 | SP9 | Sample Preparation 9 | Urine protein isolation and concentration. Service includes Isolation, partial purification, and concentration of urine proteins using ITSI Urine Protein Isolation kit. | Per sample |
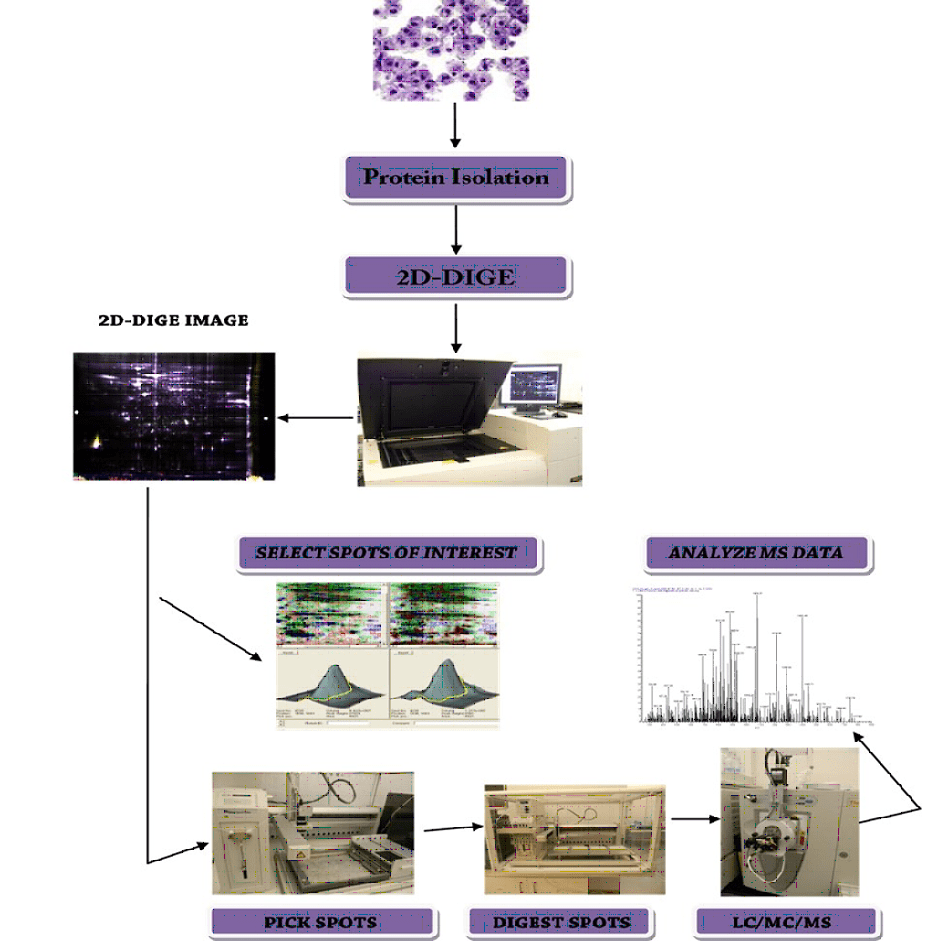
| 7 | S30 | 2D-DIGE | Two- Dimensional Difference in-Gel Electrophoresis (2D- DIGE). Quantitative and comparative proteomics using 2D-DIGE. Services include: • Experimental design, • Sample preparation to isolate total proteins using ToPI- DIGE Protein Isolation Kit, • Fractionation, quantitation and qualification of proteins, • 2D-DIGE analysis using Cy2, Cy3 and Cy5, scanning of gels using a DIGE- enabled digital scanner and • Biological variation analysis of gel images to identify differentially expressed proteins with statistically significant difference. | 2 – 10 Samples |
| 8 | S40P | Robotic spot picking | Selection and picking of fluorescent labels protein spots from 2D-DIGE gels using fully automated ETTAN spot picking robot. | 1-96 spots |
| 9 | S40D | Robotic spot digestion | In gel digestion of protein spots picked from 2D-DIGE gels using a fully automated ETTAN spot digestion robot. | 1-96 spots |
| 10 | S50.60 | LC60-MS/MS | Peptide sequencing and protein identification by liquid chromatography – tandem mass spectrometry of pure protein samples. Services include: • Tandem mass spectrometry analysis using nano-LC/MS/MS, • Database search and • Identified protein annotation – 60 min run. | Per sample |
"*" indicates required fields
Citations
Browse through a list of scientific publications and research papers involving our 2D-DIGE Services.
Identification of STAT2 serine 287 as a novel regulatory phosphorylation site in type I interferon-induced cellular responses.
H. C. Steen, S. Nogusa, R. J. Thapa, S.H. Basagoudanavar, A. Gill, S. Merali, C. A. Barrero, S. Balachandran, A. M. Gamero, The Journal of Biological Chemistry
Acute mitochondrial dysfunction after blast exposure: potential role of mitochondrial glutamate oxaloacetate transaminase.
P. Arun, R. Abu-Taleb, S. Oguntayo, Y. Wang, M. Valiyaveettil, J. Long, M.P. Nambiar Journal of Neurotrauma
Modulation of hearing related proteins in the brain and inner ear following repeated blast exposures.
P. Arun, M. Valiyaveettil1, L. Biggemann, Y. Alamneh, Y. Wei, S. Oguntayo, Y. Wang, J.B. Long, M.P. Nambiar1 … Interventional Medicine & Applied Science vol.4 2012 October 1
The effect of selenium enrichment on baker’s yeast proteome.
K. El-Bayoumy, A. Dasa, S. Russell, S. Wolfe, R. Jordan, K. Renganathan, T.P. Loughranb, R. Somiari. Journal of Proteomics-Elsevier Inc. 2011 October 21;.10.1016
Selenium-Responsive Proteins in the Sera of Selenium-Enriched Yeast-Supplemented Healthy American and Caucasian Men.
R. Sinha., I. Sinha., N. Facompreruqui., M. Elmissiry. Journal of Biomedical Science. 2009 September;35(3):559-67.
Proteomics of rat prostate lobes treated with 2-N-Hydroxylamino-1-methyl-6-phenylimidazo [4,5-b] pyridine, 5alpha-dihydrotestosterone, individually and in combination.
Boyiri, T., Somiari, RI., Russell, S., EL-Bayoumy, K. Int. J Oncol. 2009 Sep;35(3):559-67
High-throughput proteomic analysis of human infiltrating ductal carcinoma of the breast.
Somiari RI, Sullivan A, Russell S, Somiari S, Hu H, Jordan R, George A, Katenhusen R, Buchowiecka A, Arciero C, Brzeski H, Hooke J, Shriver C. Proteomics. 2003 Oct;3(10):1863-73.
Albumin depletion method for improved plasma glycoprotein analysis by two-dimensional difference gel electrophoresis.
Brzeski H, Katenhusen R, Sullivan A, Russell S, George A, Somiari RI, Shriver C. Biotechniques 2003 Dec;35(6):1128-32.
Difference In-Gel Electrophoresis in a High-Throughput Environment.
Somiari RI. Russell S, Somiari SB, Sullivan AG, Ellsworth, DL, Brzeski, H, Shriver, CD In Walker J. (Editor) The Proteomics Protocols Handbook. Humana Press Inc., NJ, USA. 2005;pp. 223 – 237.
Proteomics of breast carcinoma.
Somiari RI, Somiari S, Russell S, Shriver CD. J Chromatogr B Analyt Technol Biomed Life Sci. 2005 Feb 5;815(1-2):215-25.
Proteomics in human cancer research.
Somiari RI, Somiari S, Pastwa E, Czyz M. Proteomics. Clinical Applications. 2007 January; DOI: 10.1002/prca.200600369.

